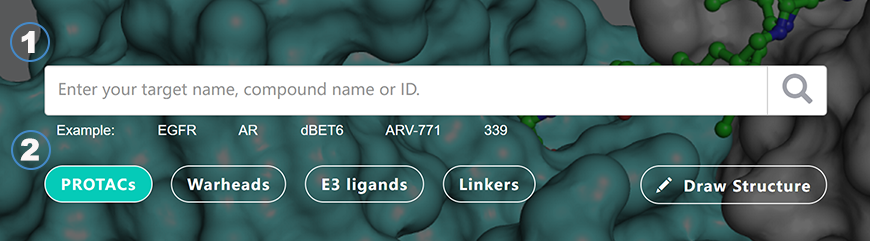
Text-based search: First, enter your target name, compound name, ID or SMILES. Then, select which dataset to search.
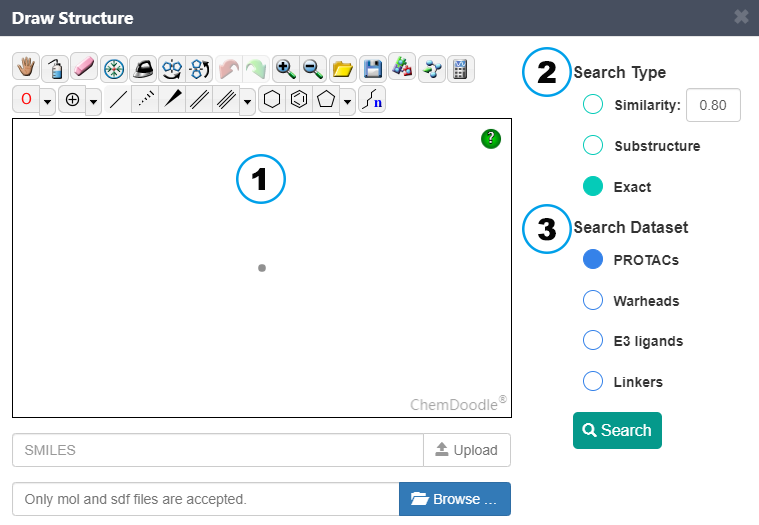
Structure-based search: First, draw your structure, input SMILES or upload a MOL/SDF file. Then, choose the search type. In the similarity search, the bit vector Morgan fingerprint, an FCFP-like fingerprint, is utilized to characterize the chemical features of a molecule. Finally, select which dataset to search.
The ternary complex structure section displays the PROTAC-mediated ternary complex. For the PROTACs with good degradation capacity but no crystal structures, we also provide the predicted ternary complex models using our PROTAC-Model method (J. Med. Chem. 2021, 64, 21, 16271-16281).
- Molecule visualization: A 3Dmol.js-enabled visualization window shows the top 10 clusters with no more than 3 representative models per cluster. Action Options: Rotate: Press and hold the left mouse button. Zoom: Use mouse's scroll button. Translate: Press and hold mouse's scroll button.
- The targeted proteins of PROTACs.
- Summary of the predicted models. In the Cluster_X_Y.pdb, X and Y represents the best rank from the same cluster and the rank of the model according to the interface energy, respectively. For example, cluster_1_4.pdb belongs to the cluster 1 and is ranked fourth based on the interface energy. PROTAC-Model is used to construct models, which combines the FRODOCK-based protocol and RosettaDock-based refinement. Top 10 clusters with no more than 3 representative models per cluster are displayed online.
- Statistics of the predicted models. Cluster: The models after clustering. Each cluster contains no more than 3 representative models. All: All models before clustering.

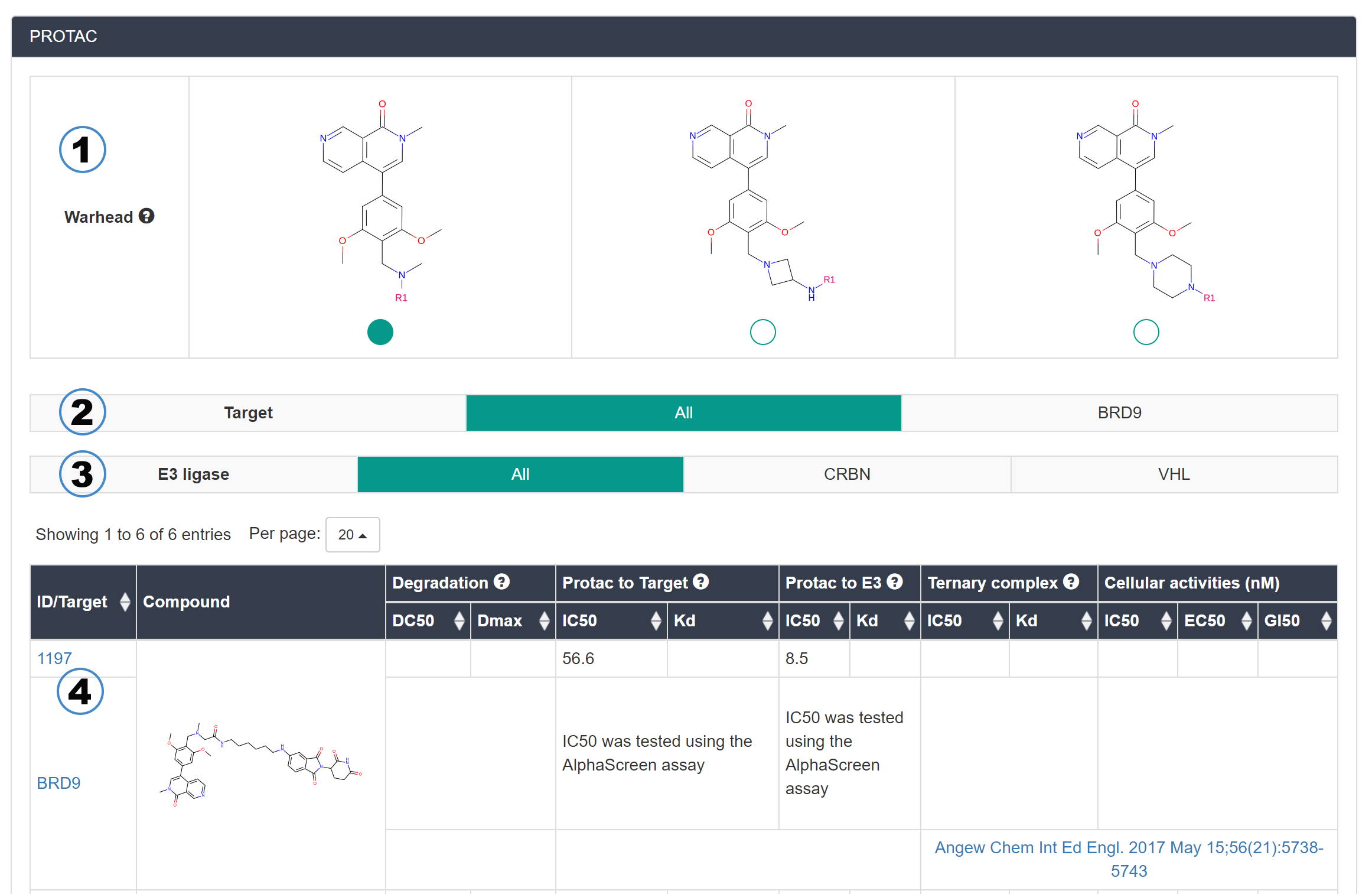
PROTACs based on this warhead or E3 ligand are also displayed on the warhead/E3 ligand page. Taking the warhead page as an example, the features are as follows.
- The warheads based on same initial structures: The initial structures of warheads might be modified for integration into PROTACs. Thus, this part summarizes these structures to facilitate users to retrieve.
- The targeted proteins of PROTACs.
- The E3 ligases of PROTACs (Notice: E3 ligand page doesn't have this filtering option).
- The list of PROTACs.