Introduction
Protein-protein interactions (PPIs) have been regarded as an attractive emerging class of therapeutic targets for the development of new treatments. Computational approaches, especially molecular docking, have been extensively employed to predict the binding structures of PPI-inhibitors or discover novel small molecule PPI inhibitors. However, due to the relatively ‘undruggable’ features of PPI interfaces, accurate predictions of the binding structures for ligands towards PPI targets are quite challenging for most docking algorithms. Here, we constructed a non-redundant pose ranking benchmark dataset for small-molecule PPI inhibitors which contains 900 binding poses for 184 protein-ligand complexes. Then, we evaluated the performance of MM/PB(GB)SA to identify the correct binding poses for PPI inhibitors, including two Prime MM/GBSA procedures from the Schrödinger suite and seven different MM/PB(GB)SA procedures from the Amber package. Our results showed that MM/PBSA outperformed the Glide SP scoring function (success rate of 58.6%) and MM/GBSA in most cases, especially the PB3 procedure which can achieve an overall success rate about 74%. Moreover, the GB6 procedure (success rate of 68.9%) performs much better than the other MM/GBSA procedures, highlighting the excellent potential of the GBNSR6 implicit solvation model for pose ranking. Finally, we developed the webserver of Fast Amber Rescoring for PPI Inhibitors (http://cadd.zju.edu.cn/farppi/), which offers a freely available service for rescoring the docking poses for PPI inhibitors by using the MM/PB(GB)SA methods.
If you use farPPI, please cite: Zhe Wang, Xuwen Wang, Youyong Li, Tailong Lei, Ercheng Wang, Dan Li, Yu Kang, Feng Zhu, Tingjun Hou; farPPI: a webserver for accurate prediction of protein-ligand binding structures for small-molecule PPI inhibitors by MM/PB(GB)SA methods, Bioinformatics, 2019, 35, 1777–1779
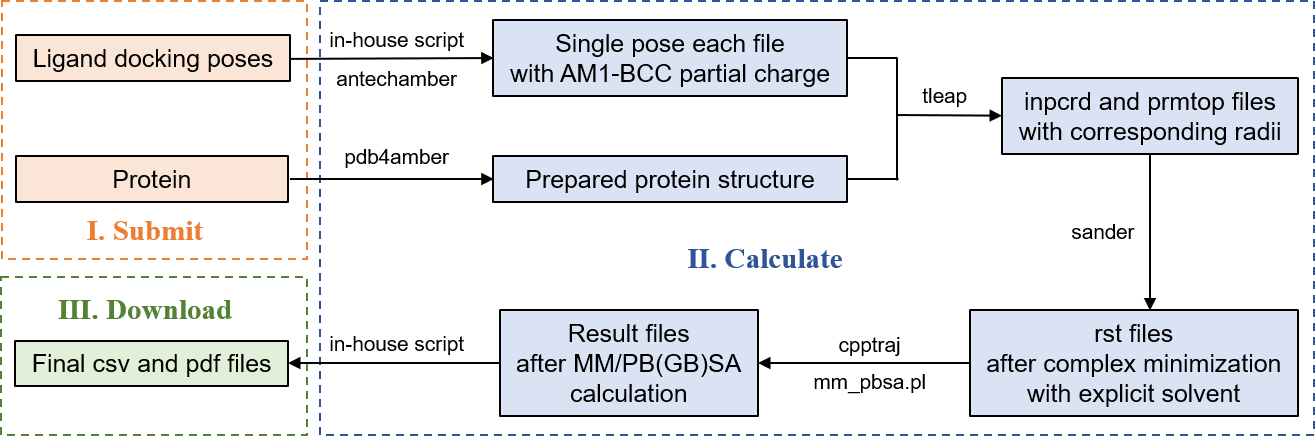
Workflow
Server status
4736
Finished jobs
163
Failed/killed jobs
0
Running jobs
0