User's manual
The procedure for using DeepChargePredictor server can be roughly divided into 6 steps as shown in Figure, including:
- Name the project.
- Supply an email address for returning the prediction result.
- Choose the prediction method.
- Choose the type of charge for prediction.
- Choose the format of the molecule file for upload (mol2/sdf).
- Choose the molecule file (one/multiple in a single file).
- Submit the job and/or wait for the result online (depending on whether an email is supplied).
Workflow of DeepChargePredictor server
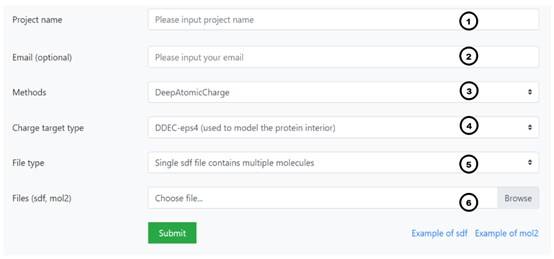
Name your project at position 1. The length of the project name should be limited to 3~20 characters.
Email is optional as shown in position 2. If the user does not provide the email address, he/she may need to wait for the result on the page. After processing the uploaded file in the backend, the browser will directly download the calculation result (mol2 file), which is very convenient for small tasks such as the prediction for a single molecule. Whereas, the user is recommended to fill the email address when calculating a large number of molecules since he/she may need to wait for a long time on the page. With the email supplied, a download link will be sent to the email address as soon as the calculation is complete.
The prediction method can be chosen at position 3, where two methods, including APD and DAC, were incorporated. However, in most cases, we recommend the DAC method since it runs faster and does not need any optimization of the input molecules prior to submit. As shown in the benchmark part of the paper, the difference between the predicted and the original charges will not significantly affect the prediction result in practice.
For the charge type, the server provides four kinds of charges, namely RESP, AM1-BCC, DDEC-eps4 and DDEC-eps78 charges. The type of charge for predicting can be chosen at position 4. Among them, the RESP and DDEC charges were calculated based on high-level QM method, while the AM1-BCC charges were calculated based on semi-empirical method.
Choose the format of the upload file. DeepChargePredictor server supports the input format of sdf and mol2 files. In this option, as shown in position 5, the user can upload a single file containing one or multiple molecules for the calculation.
When uploading multiple files, the user can select multiple files directly without compression. Due to the limited computing resources on the server, we recommend the maximum number of the upload molecules to 10000 for DAC. Whereas, since the APD algorithm works much slower than the APD algorithm, we recommend the maximum number to 2000 for the APD-based methods. Position 6 shows the file number that the user is ready to submit. If the maximum number of the upload files is exceeded, a warning will be returned.
The validation process consists of two parts: the validation of the file format and the validation of the content. Here, the so-called valid format means that the submitted mol2 or sdf file can be recognized by RDKit and OpenBabel. A second validation is used to check whether the molecules submitted by the users contain atom types beyond those in the training set of the models. Specifically, the DAC models contain 10 elements, including C, H, N, O, S, P, F, Cl, Br, and I. Thus, if the files submitted by the users contain some atoms beyond these 10 elements, they will be recognized as the other elements, which will lead to unreasonable prediction result and should be used with caution. Since the APD method is designed based on the SYBYL type elements, it is necessary to ensure that the types of the atoms in the input molecules are within the 25 SYBYL types, including H, C.cat, C.ar, C.2, C.3, C.1, N.3, N.2, N.1, N.ar, N.am, N.pl3, N.4, O.3, O.2, O.co2, P.3, S.3, S.2, S.O, S.O2, F, Cl, Br and I. If the input file fails in the validation process, the backend will ignore the wrong file and an error message will be returned.