Instructions
DeepChargePredictor is a server to predict QM-based atomic charges for small molecules, including RESP, AM1-BCC and DDEC charges, which are widely used in biological system simulations, such as structure-based drug design. Currently, the server contains three state-of-the-art machine-learning algorithms developed by Hou group, namely, the Atom-Path-Descriptor (APD), the DeepAtomicCharge (DAC) and SuperAtomicCharge (SAC).
APD algorithm
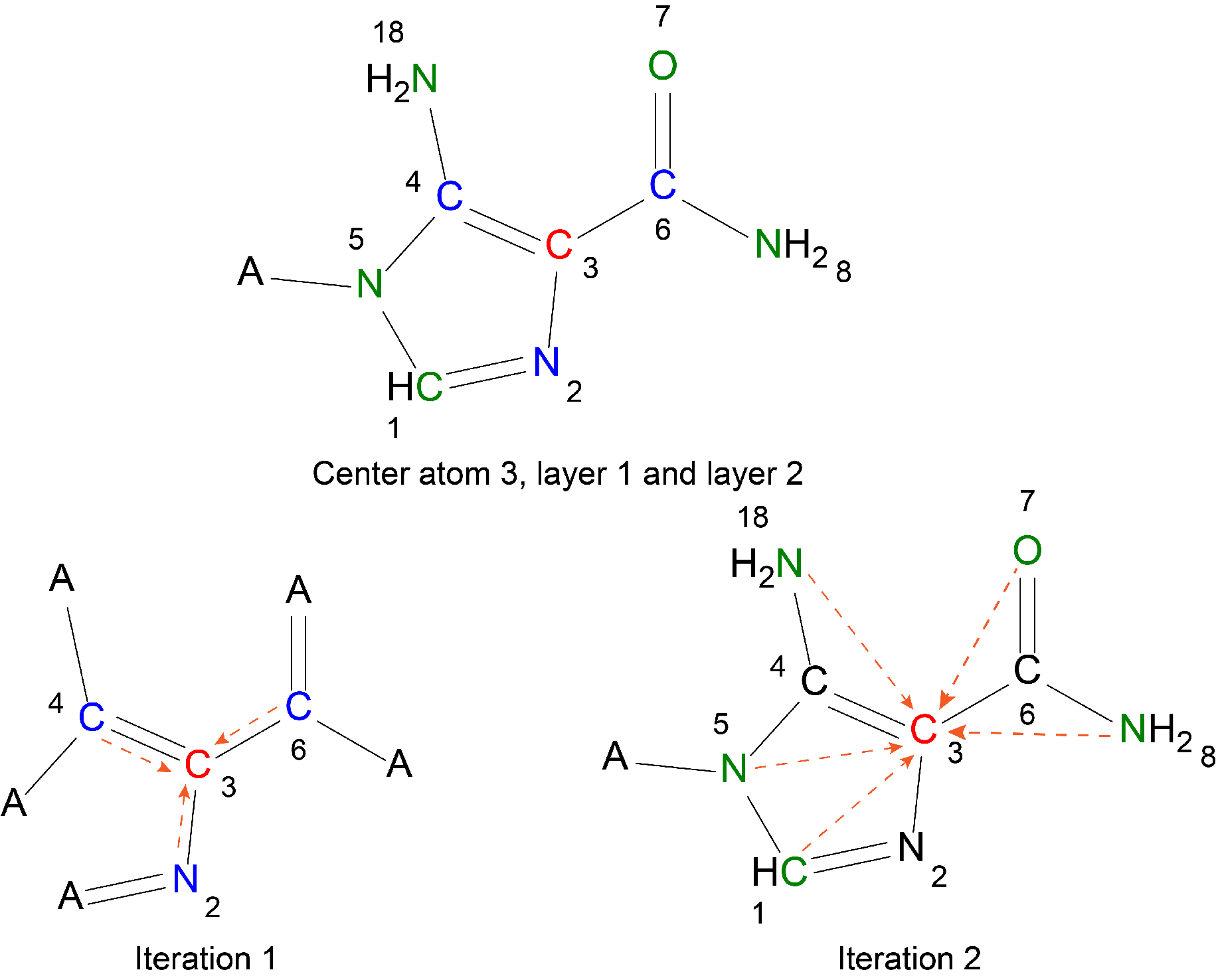
The APD algorithm was constructed in the combination of XGBoost, in which the atomic centers and atomic layers were assigned to the 3D structure of the molecules based on the Atom-Pair-Paths to characterize the local chemical environment.
DAC algorithm
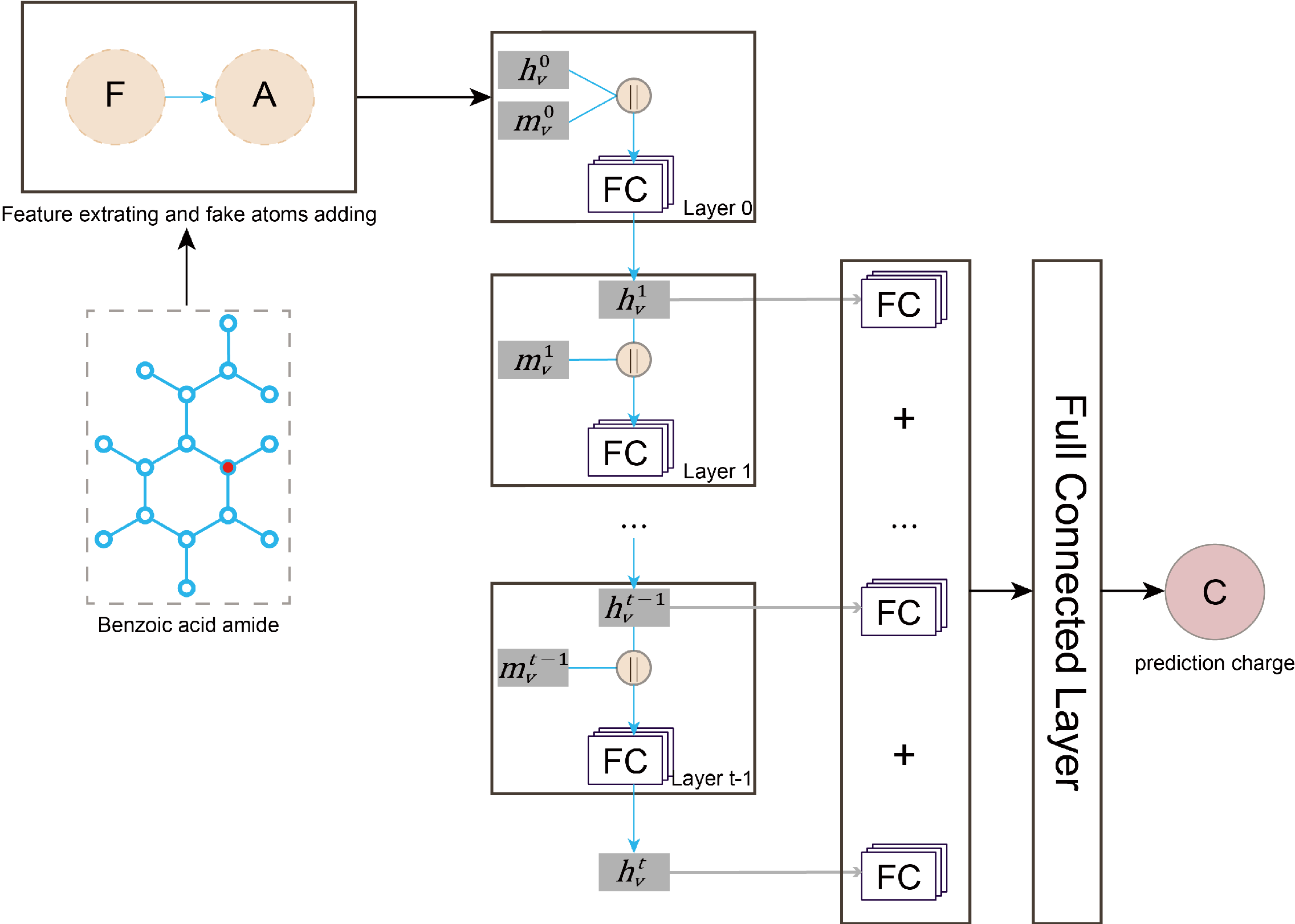
The DAC models were constructed based on the graph convolution network (GCN), which can dynamically learn the appropriate features from the basic atomic properties and connection information of the given molecules without any prior knowledge of them.
SAC algorithm
The SAC models were designed to simultaneously exploit the 2D and 3D structural information of molecule structures based on the graph convolution network (GCN).
