N-[3-[5-ethyl-2-[2-methoxy-4-(4-methylpiperazin-1-yl)anilino]-6,7-dioxo-pteridin-8-yl]phenyl]prop-2-enamide
Inhibitor information
- CovInDB Inhibitor
- CI004943
- Name
- N-[3-[5-ethyl-2-[2-methoxy-4-(4-methylpiperazin-1-yl)anilino]-6,7-dioxo-pteridin-8-yl]phenyl]prop-2-enamide
- Molecular Formula
- C29H32N8O4
- Molecular Weight
- 556.2546515 g/mol
- Structure
-
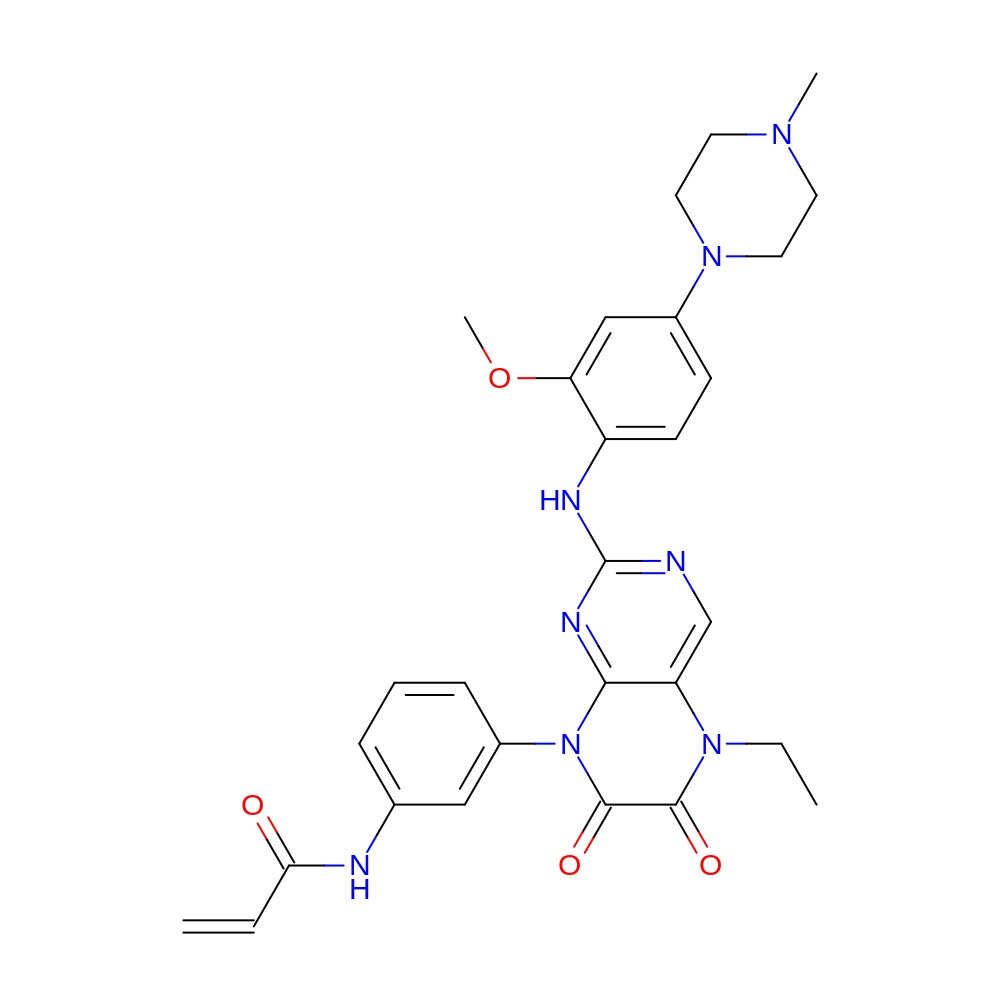
- IUPAC Name
- N-[3-[5-ethyl-2-[2-methoxy-4-(4-methylpiperazin-1-yl)anilino]-6,7-dioxo-pteridin-8-yl]phenyl]prop-2-enamide
- InChI
- InChI=1S/C29H32N8O4/c1-5-25(38)31-19-8-7-9-21(16-19)37-26-23(36(6-2)27(39)28(37)40)18-30-29(33-26)32-22-11-10-20(17-24(22)41-4)35-14-12-34(3)13-15-35/h5,7-11,16-18H,1,6,12-15H2,2-4H3,(H,31,38)(H,30,32,33)
- InChI Key
- LKQWOHYTZVPINL-UHFFFAOYSA-N
- Canonical SMILES
- C=CC(=O)Nc1cccc(-n2c(=O)c(=O)n(CC)c3cnc(Nc4ccc(N5CCN(C)CC5)cc4OC)nc32)c1
- Cocrystal structures
- No cocrystal structures found for this inhibitor.
Calculated Properties
- Molecular Weight
-
556.2546515 g/mol
Computed by RDKit
- logP
-
2.627
Computed by ALOGPS
- logS
-
-5.039
Computed by ALOGPS
- Heavy Atom Count
-
41
Computed by RDKit
- Ring Count
-
5
Computed by RDKit
- Hydrogen Bond Acceptor Count
-
11
Computed by RDKit
- Hydrogen Bond Donor Count
-
2
Computed by RDKit
- Rotatable Bond Count
-
8
Computed by RDKit
- Topological Polar Surface Area
-
126.62 Å2
Computed by RDKit
3D Structure
targets
| Name | ID | Warhead | Reaction Mechanism | Target Site | Activity Type | Relation | Value | Unit | Experiment Method | Assay | Reference |
|---|
bioactivity
| Object | Object Type | Activity Type | Relation | Value | Unit | Assay | Reference |
|---|
Similar compounds in Virtual Screening library
Similar Natural compounds
No similar natural compounds found for this inhibitor.