4FWD
Target information
- RCSB PDB
- 4FWD
- Title
- Crystal structure of the Lon-like protease MtaLonC in complex with bortezomib
- Method
- X-RAY DIFFRACTION
- Resolution
- 2.03
- Classification
- HYDROLASE/HYDROLASE INHIBITOR
- Organism
- Meiothermus taiwanensis
- Protein
- Endopeptidase La (C9DRU9)
- Year
- 2012
- Publication Title
- Structures of an ATP-independent Lon-like protease and its complexes with covalent inhibitors
- Abstract
-
The Lon proteases are a unique family of chambered proteases with a built-in AAA+ (ATPases associated with diverse cellular activities) module. Here, crystal structures of a unique member of the Lon family with no intrinsic ATPase activity in the proteolytically active form are reported both alone and in complexes with three covalent inhibitors: two peptidomimetics and one derived from a natural product. This work reveals the unique architectural features of an ATP-independent Lon that selectively degrades unfolded protein substrates. Importantly, these results provide mechanistic insights into the recognition of inhibitors and polypeptide substrates within the conserved proteolytic chamber, which may aid the development of specific Lon-protease inhibitors.
- External Link
- RCSB PDB
Ligand information
- HET
- BO2
- Chain ID
- A
- HET Number
- 801
- Molecular Formula
- C19H25BN4O4
- Structure
-
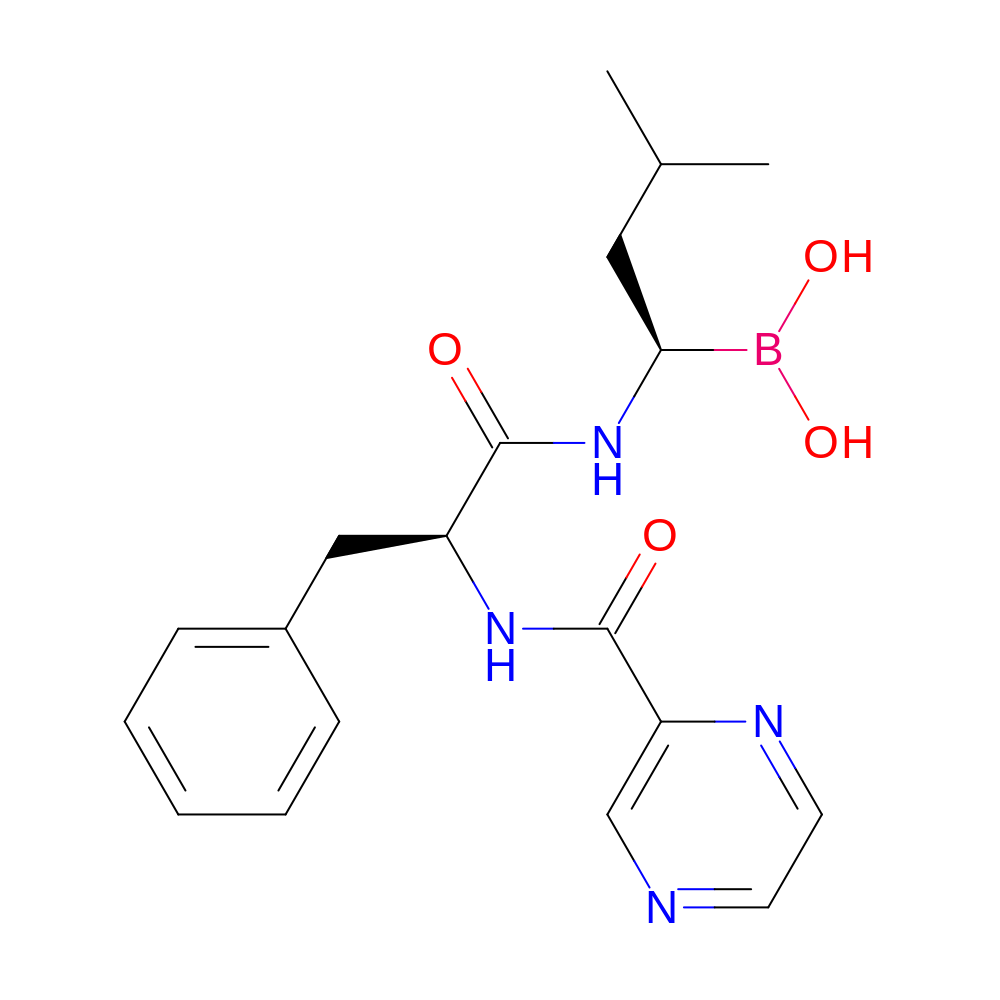
- IUPAC Name
- [(1R)-3-methyl-1-[[(2S)-3-phenyl-2-(pyrazine-2-carbonylamino)propanoyl]amino]butyl]boronic acid
- InChI
- InChI=1S/C19H25BN4O4/c1-13(2)10-17(20(27)28)24-18(25)15(11-14-6-4-3-5-7-14)23-19(26)16-12-21-8-9-22-16/h3-9,12-13,15,17,27-28H,10-11H2,1-2H3,(H,23,26)(H,24,25)/t15-,17-/m0/s1
- InChI Key
- GXJABQQUPOEUTA-RDJZCZTQSA-N
- Canonical SMILES
- c1ccccc1C[C@@H](C(=O)N[C@H](B(O)O)CC(C)C)NC(=O)c2cnccn2
- Bioactivity data
- CI006008
Covalent Binding
- Warhead
- Boronic Acid
- Reaction Mechanism
- Boronic Acid Addition
- Residue
- SER : 582
- Residue Chain
- A
- Interactions
- Pharmacophore Model